JupyterHub: Difference between revisions
No edit summary |
|||
| (108 intermediate revisions by 10 users not shown) | |||
| Line 1: | Line 1: | ||
<languages /> | |||
<translate> | |||
<!--T:2--> | |||
<i>JupyterHub is the best way to serve Jupyter Notebook for multiple users. It can be used in a class of students, a corporate data science group or scientific research group.</i> | |||
<ref>http://jupyterhub.readthedocs.io/en/latest/index.html</ref> | |||
JupyterHub | <!--T:3--> | ||
JupyterHub provides a preconfigured version of JupyterLab and/or Jupyter Notebook; for more configuration options, please check the [[Jupyter]] page. | |||
{{Warning | |||
|title=Running notebooks | |||
|content=Jupyter Lab and notebooks are meant for '''short''' interactive tasks such as testing, debugging or quickly visualize data (few minutes). Running longer analysis must be done in an [[Running_jobs#Use_sbatch_to_submit_jobs|non-interactive job (sbatch)]]. | |||
See also [[JupyterHub#Running_notebooks_as_Python_scripts|how to run notebooks as python scripts below]]. | |||
}} | |||
= Alliance initiatives = <!--T:4--> | |||
<!--T:5--> | |||
Some regional initiatives offer access to computing resources through JupyterHub. | |||
== JupyterHub on clusters == <!--T:11--> | |||
<!--T:8--> | |||
On the following clusters[[#clusters_note|<sup>‡</sup>]], use your Alliance username and password to connect to JupyterHub: | |||
{| class="wikitable" | |||
|- | |||
! JupyterHub !! Comments | |||
|- | |||
| <b>[https://jupyterhub.beluga.alliancecan.ca/ Béluga]</b> || Provides access to JupyterLab servers spawned through jobs on the [[Béluga/en|Béluga]] cluster. | |||
|- | |||
| <b>[https://jupyterhub.cedar.computecanada.ca/ Cedar]</b> || Provides access to JupyterLab servers spawned through jobs on the [[Cedar]] cluster. | |||
|- | |||
| <b>[https://jupyterhub.narval.alliancecan.ca/ Narval]</b> || Provides access to JupyterLab servers spawned through jobs on the [[Narval/en|Narval]] cluster. | |||
|- | |||
| <b>[https://jupyter.scinet.utoronto.ca/ Niagara]</b> || This is a node which has been designated as a Jupyter Hub and it can run Jupyter Notebook sessions. To learn more, see the [https://docs.scinet.utoronto.ca/index.php/Jupyter_Hub SciNet JupyterHub wiki page]. | |||
|- | |||
| <b>[https://jupyterhub.sharcnet.ca/ Graham]</b> || Provides access to JupyterLab servers spawned through jobs on the [[Graham]] cluster. | |||
|} | |||
== References = | <!--T:18--> | ||
<b><sup id="clusters_note">‡</sup> Note that the compute nodes running the Jupyter kernels do not have internet access</b>. This means that you can only transfer files from/to your own computer; you cannot download code or data from the internet (e.g. cannot do "git clone", cannot do "pip install" if the wheel is absent from our [[Available Python wheels|wheelhouse]]). You may also have problems if your code performs downloads or uploads (e.g. in machine learning where downloading data from the code is often done). | |||
== JupyterHub for universities and schools == <!--T:12--> | |||
<!--T:6--> | |||
* The [https://www.pims.math.ca Pacific Institute for the Mathematical Sciences] in collaboration with the Alliance and [http://www.cybera.ca Cybera] offer cloud-based hubs to universities and schools. Each institution can have its own hub where users authenticate with their credentials from that institution. The hubs are hosted on Alliance [[Cloud|clouds]] and are essentially for training purposes. Institutions interested in obtaining their own hub can visit [http://syzygy.ca Syzygy]. | |||
= Server options = <!--T:13--> | |||
<!--T:14--> | |||
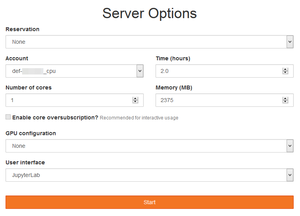
[[File:JupyterHub_Server_Options.png|thumb|<i>Server Options</i> form on Béluga's JupyterHub]] | |||
Once logged in, depending on the configuration of JupyterHub, the user's web browser is redirected to either | |||
<b>a)</b> a previously launched Jupyter server, | |||
<b>b)</b> a new Jupyter server with default options, or | |||
<b>c)</b> a form that allows a user to set different options for their Jupyter server before pressing the <i>Start</i> button. | |||
In all cases, it is equivalent to accessing requested resources via an [[Running_jobs#Interactive_jobs|interactive job]] on the corresponding cluster. | |||
<!--T:64--> | |||
<b>Important:</b> On each cluster, only one interactive job at a time gets a priority increase in order to start in a few seconds or minutes. That includes <code>salloc</code>, <code>srun</code> and JupyterHub jobs. If you already have another interactive job running on the cluster hosting JupyterHub, your new Jupyter session may never start before the time limit of 5 minutes. | |||
== Compute resources == <!--T:15--> | |||
<!--T:16--> | |||
For example, <i>Server Options</i> available on [https://jupyterhub.beluga.computecanada.ca/ Béluga's JupyterHub] are: | |||
* <i>Account</i> to be used: any <code>def-*</code>, <code>rrg-*</code>, <code>rpp-*</code> or <code>ctb-*</code> account a user has access to | |||
* <i>Time (hours)</i> required for the session | |||
* <i>Number of (CPU) cores</i> that will be reserved on a single node | |||
* <i>Memory (MB)</i> limit for the entire session | |||
* (Optional) <i>GPU configuration</i>: at least one GPU | |||
* <i>[[JupyterHub#User_Interface|User interface]]</i> (see below) | |||
== User interface == <!--T:9--> | |||
<!--T:17--> | |||
While JupyterHub allows each user to use one Jupyter server at a time on each hub, there can be multiple options under <i>User interface</i>: | |||
* Jupyter Notebook (classic interface): Even though it offers many functionalities, the community is moving towards [[JupyterHub#JupyterLab|JupyterLab]], which is a better platform that offers many more features. | |||
* <b>[[JupyterHub#JupyterLab|JupyterLab]]</b> (modern interface): This is the most recommended Jupyter user interface for interactive prototyping and data visualization. | |||
* Terminal (for a single terminal only): It gives access to a terminal connected to a remote account, which is comparable to connecting to a server through an SSH connection. | |||
<!--T:10--> | |||
Note: JupyterHub could also have been configured to force a specific user interface. This is usually done for special events. | |||
= JupyterLab = <!--T:19--> | |||
<!--T:20--> | |||
JupyterLab is now the recommended general-purpose user interface to use on a JupyterHub. | |||
From a JupyterLab server, you can manage your remote files and folders, | |||
and you can launch Jupyter applications like a terminal, (Python 3) notebooks, RStudio and a Linux desktop. | |||
== The JupyterLab interface == <!--T:21--> | |||
<!--T:22--> | |||
When JupyterLab is ready to be used, the interface has multiple panels. | |||
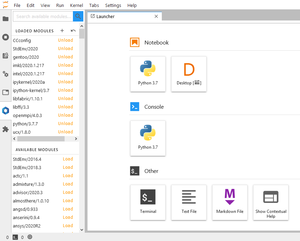
[[File:JupyterLab_HomeTab.png|thumb|Default home tab when JupyterLab is loaded]] | |||
=== Menu bar on top === <!--T:23--> | |||
<!--T:24--> | |||
* In the <i>File</i> menu: | |||
** <i>Hub Control Panel</i>: if you want to manually stop the JupyterLab server and the corresponding job on the cluster. This is useful when you want to start a new JupyterLab server with more or less resources. | |||
** <i>Log Out</i>: the JupyterHub session will end, which will also stop the JupyterLab server and the corresponding job on the cluster. | |||
* Most other menu items are related to notebooks and Jupyter applications. | |||
=== Tool selector on left === <!--T:25--> | |||
<!--T:26--> | |||
* <i>File Browser</i> (folder icon): | |||
** This is where you can browse in your home, project and scratch spaces. | |||
** It is also possible to upload files. | |||
* <i>Running Terminals and Kernels</i> (stop icon): | |||
** To stop kernel sessions and terminal sessions | |||
* <i>Commands</i> | |||
* <i>Property Inspector</i> | |||
* <i>Open Tabs</i>: | |||
** To navigate between application tabs. | |||
** To close application tabs (the corresponding kernels remain active). | |||
[[File:JupyterLab_Softwares.png|thumb|Loaded modules and available modules]] | |||
* <b><i>Software</i></b> (blue diamond sign): | |||
** Alliance modules can be loaded and unloaded in the JupyterLab session. Depending on the modules loaded, icons directing to the corresponding [[#Prebuilt_applications|Jupyter applications]] will appear in the <i>Launcher</i> tab. | |||
** The search box can search for any [[Available software|available module]] and show the result in the <i>Available Modules</i> subpanel. Note: Some modules are hidden until their dependency is loaded: we recommend that you first look for a specific module with <code>module spider module_name</code> from a terminal. | |||
** The next subpanel is the list of <i>Loaded Modules</i> in the whole JupyterLab session. Note: While <code>python</code> and <code>ipython-kernel</code> modules are loaded by default, additional modules must be loaded before launching some other applications or notebooks. For example: <code>scipy-stack</code>. | |||
** The last subpanel is the list of <i>Available modules</i>, similar to the output of <code>module avail</code>. By clicking on a module's name, detailed information about the module is displayed. By clicking on the <i>Load</i> link, the module will be loaded and added to the <i>Loaded Modules</i> list. | |||
=== Applications area on right === <!--T:27--> | |||
<!--T:28--> | |||
* The <i>Launcher</i> tab is open by default. | |||
** It contains all available [[#Prebuilt_applications|Jupyter applications and notebooks]], depending on which modules are loaded | |||
=== Status bar at the bottom === <!--T:29--> | |||
<!--T:30--> | |||
* By clicking on the icons, this brings you to the <i>Running Terminals and Kernels</i> tool. | |||
== Prebuilt applications == <!--T:31--> | |||
<!--T:32--> | |||
JupyterLab offers access to a terminal, an IDE (Desktop), a Python console and different options to create text and markdown files. This section presents only the main supported Jupyter applications that work with our software stack. | |||
=== Command line interpreters === <!--T:33--> | |||
<!--T:34--> | |||
[[File:JupyterLab_Launcher_Julia.png|thumb|Julia console launcher button]] | |||
[[File:JupyterLab_Launcher_Python.png|thumb|Python console launcher button]] | |||
[[File:JupyterLab_Launcher_Terminal.png|thumb|Terminal launcher button]] | |||
==== Julia console ==== <!--T:35--> | |||
<!--T:36--> | |||
To enable the <i>Julia 1.x</i> console launcher, an <code>ijulia-kernel</code> module needs to be loaded. When launched, a Julia interpreter is presented in a new JupyterLab tab. | |||
==== Python console ==== <!--T:37--> | |||
<!--T:38--> | |||
The <i>Python 3.x</i> console launcher is available by default in a new JupyterLab session. When launched, a Python 3 interpreter is presented in a new JupyterLab tab. | |||
==== Terminal ==== <!--T:39--> | |||
<!--T:40--> | |||
This application launcher will open a terminal in a new JupyterLab tab: | |||
* The terminal runs a (Bash) shell on the remote compute node without the need of an SSH connection. | |||
** Gives access to the remote filesystems (<code>/home</code>, <code>/project</code>, <code>/scratch</code>). | |||
** Allows running compute tasks. | |||
* The terminal allows copy-and-paste operations of text: | |||
** Copy operation: select the text, then press Ctrl+C. | |||
*** Note: Usually, Ctrl+C is used to send a SIGINT signal to a running process, or to cancel the current command. To get this behaviour in JupyterLab's terminal, click on the terminal to deselect any text before pressing Ctrl+C. | |||
** Paste operation: press Ctrl+V. | |||
=== Available notebook kernels === <!--T:41--> | |||
==== Julia notebook ==== <!--T:42--> | |||
<!--T:43--> | |||
To enable the <i>Julia 1.x</i> notebook launcher, an <code>ijulia-kernel</code> module needs to be loaded. When launched, a Julia notebook is presented in a new JupyterLab tab. | |||
==== Python notebook ==== <!--T:44--> | |||
<!--T:45--> | |||
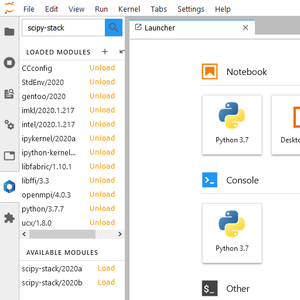
[[File:JupyterLab_Softwares_ScipyStack.png|thumb|Searching for scipy-stack modules]] | |||
If any of the following scientific Python packages is required by your notebook, before you open this notebook, you must load the <code>scipy-stack</code> module from the JupyterLab <i>Softwares</i> tool: | |||
* <code>ipython</code>, <code>ipython_genutils</code>, <code>ipykernel</code>, <code>ipyparallel</code> | |||
* <code>matplotlib</code> | |||
* <code>numpy</code> | |||
* <code>pandas</code> | |||
* <code>scipy</code> | |||
* Other notable packages are <code>Cycler</code>, <code>futures</code>, <code>jupyter_client</code>, <code>jupyter_core</code>, <code>mpmath</code>, <code>pathlib2</code>, <code>pexpect</code>, <code>pickleshare</code>, <code>ptyprocess</code>, <code>pyzmq</code>, <code>simplegeneric</code>, <code>sympy</code>, <code>tornado</code>, <code>traitlets</code>. | |||
* And many more (click on the <code>scipy-stack</code> module to see all <i>Included extensions</i>). | |||
<!--T:46--> | |||
Note: You may also install needed packages by running for example the following command inside a cell: <code>!pip install --no-index numpy</code>. | |||
* For some packages (like <code>plotly</code>, for example), you may need to restart the notebook's kernel before importing the package. | |||
* The installation of packages in the default Python kernel environment is temporary to the lifetime of the JupyterLab session; you will have to reinstall these packages the next time you start a new JupyterLab session. For a persistent Python environment, you must configure a <b>[[Advanced_Jupyter_configuration#Python_kernel|custom Python kernel]]</b>. | |||
<!--T:47--> | |||
To open an existing Python notebook: | |||
* Go back to the <i>File Browser</i>. | |||
* Browse to the location of the <code>*.ipynb</code> file. | |||
* Double-click on the <code>*.ipynb</code> file. | |||
** This will open the Python notebook in a new JupyterLab tab. | |||
** An IPython kernel will start running in the background for this notebook. | |||
<!--T:48--> | |||
To open a new Python notebook in the current <i>File Browser</i> directory: | |||
* Click on the <i>Python 3.x</i> launcher under the <i>Notebook</i> section. | |||
** This will open a new Python 3 notebook in a new JupyterLab tab. | |||
** A new IPython kernel will start running in the background for this notebook. | |||
=== Other applications === <!--T:49--> | |||
==== OpenRefine ==== <!--T:50--> | |||
<!--T:51--> | |||
[[File:JupyterLab_Launcher_OpenRefine.png|thumb|OpenRefine launcher button]] | |||
To enable the <i>OpenRefine</i> application launcher, an <code>openrefine</code> module needs to be loaded. Depending on the software environment version, the latest version of OpenRefine should be loaded: | |||
* with <code>StdEnv/2023</code>, no OpenRefine module is available as of August 2024; please load <code>StdEnv/2020</code> first. | |||
* with <code>StdEnv/2020</code>, load module: <code>openrefine/3.4.1</code>. | |||
<!--T:52--> | |||
This <i>OpenRefine</i> launcher will open or reopen an OpenRefine interface in a new web browser tab. | |||
* It is possible to reopen an active OpenRefine session after the web browser tab was closed. | |||
* The OpenRefine session will end when the JupyterLab session ends. | |||
==== RStudio ==== <!--T:53--> | |||
<!--T:54--> | |||
[[File:JupyterLab_Launcher_RStudio.png|thumb|RStudio launcher button]] | |||
To enable the <i>RStudio</i> application launcher, load the module: <code>rstudio-server/4.3</code> | |||
<!--T:56--> | |||
This <i>RStudio</i> launcher will open or reopen an RStudio interface in a new web browser tab. | |||
* It is possible to reopen an active RStudio session after the web browser tab was closed. | |||
* The RStudio session will end when the JupyterLab session ends. | |||
==== VS Code ==== <!--T:57--> | |||
<!--T:58--> | |||
[[File:JupyterLab_Launcher_VSCode.png|thumb|VS Code launcher button]] | |||
To enable the <i>VS Code</i> (Visual Studio Code) application launcher, a <code>code-server</code> module needs to be loaded. Depending on the software environment version, the latest version of VS Code should be loaded: | |||
* with <code>StdEnv/2023</code>, load module: <code>code-server/4.92.2</code>. | |||
* with <code>StdEnv/2020</code>, load module: <code>code-server/3.12.0</code>. | |||
<!--T:59--> | |||
This <i>VS Code</i> launcher will open or reopen the VS Code interface in a new web browser tab. | |||
* For a new session, the <i>VS Code</i> session can take up to 3 minutes to complete its startup. | |||
* It is possible to reopen an active VS Code session after the web browser tab was closed. | |||
* The VS Code session will end when the JupyterLab session ends. | |||
==== Desktop ==== <!--T:60--> | |||
<!--T:61--> | |||
[[File:JupyterLab_Launcher_Desktop.png|thumb|Desktop launcher button]] | |||
This <i>Desktop</i> launcher will open or reopen a remote Linux desktop interface in a new web browser tab: | |||
* This is equivalent to running a [[VNC#Compute_Nodes|VNC server on a compute node]], then creating an [[SSH_tunnelling|SSH tunnel]] and finally using a [[VNC#Setup|VNC client]], but you need nothing of all this with JupyterLab! | |||
* It is possible to reopen an active desktop session after the web browser tab was closed. | |||
* The desktop session will end when the JupyterLab session ends. | |||
== Running notebooks as Python scripts == <!--T:65--> | |||
<!--T:66--> | |||
1. From the console, or in a new notebook cell, install <tt>nbconvert</tt> : | |||
<syntaxhighlight lang="bash">!pip install --no-index nbconvert</syntaxhighlight> | |||
<!--T:67--> | |||
2. Convert your notebooks to Python scripts | |||
<syntaxhighlight lang="bash">!jupyter nbconvert --to python my-current-notebook.ipynb</syntaxhighlight> | |||
<!--T:68--> | |||
3. Create your [[Running_jobs#Use_sbatch_to_submit_jobs|non-interactive submission script]], and submit it. | |||
<!--T:69--> | |||
In your submission script, run your converted notebook with: | |||
<syntaxhighlight lang="bash">python my-current-notebook.py</syntaxhighlight> | |||
<!--T:70--> | |||
And submit your non-interactive job: | |||
{{Command | |||
|sbatch my-submit.sh | |||
}} | |||
= Possible error messages = <!--T:62--> | |||
<!--T:63--> | |||
Most JupyterHub errors are caused by the underlying job scheduler which is either unresponsive or not able to find appropriate resources for your session. For example: | |||
<!--T:71--> | |||
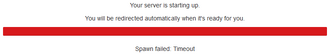
<b>Spawn failed: Timeout</b> | |||
[[File:JupyterHub Spawn failed Timeout.png|thumb|upright=1.1|JupyterHub - Spawn failed: Timeout]] | |||
* When starting a new session, JupyterHub automatically submits on your behalf a new [[Running_jobs#Interactive_jobs|interactive job]] to the cluster. If the job does not start within five minutes, a "Timeout" error message is raised and the session is cancelled. | |||
** Just like any interactive job on any cluster, a longer requested time can cause a longer wait time in the queue. Requesting a GPU or too many CPU cores can also cause a longer wait time. Make sure to request only the resources you need for your session. | |||
** If you already have another interactive job on the same cluster, your Jupyter session will be waiting along with other regular batch jobs in the queue. If possible, stop or cancel any other interactive job before using JupyterHub. | |||
** There may be just no resource available at the moment. Check the [https://status.alliancecan.ca/ status page] for any issue and try again later. | |||
= References = <!--T:7--> | |||
</translate> | |||
Latest revision as of 14:50, 8 November 2024
JupyterHub is the best way to serve Jupyter Notebook for multiple users. It can be used in a class of students, a corporate data science group or scientific research group. [1]
JupyterHub provides a preconfigured version of JupyterLab and/or Jupyter Notebook; for more configuration options, please check the Jupyter page.
Jupyter Lab and notebooks are meant for short interactive tasks such as testing, debugging or quickly visualize data (few minutes). Running longer analysis must be done in an non-interactive job (sbatch). See also how to run notebooks as python scripts below.
Alliance initiatives
Some regional initiatives offer access to computing resources through JupyterHub.
JupyterHub on clusters
On the following clusters‡, use your Alliance username and password to connect to JupyterHub:
| JupyterHub | Comments |
|---|---|
| Béluga | Provides access to JupyterLab servers spawned through jobs on the Béluga cluster. |
| Cedar | Provides access to JupyterLab servers spawned through jobs on the Cedar cluster. |
| Narval | Provides access to JupyterLab servers spawned through jobs on the Narval cluster. |
| Niagara | This is a node which has been designated as a Jupyter Hub and it can run Jupyter Notebook sessions. To learn more, see the SciNet JupyterHub wiki page. |
| Graham | Provides access to JupyterLab servers spawned through jobs on the Graham cluster. |
‡ Note that the compute nodes running the Jupyter kernels do not have internet access. This means that you can only transfer files from/to your own computer; you cannot download code or data from the internet (e.g. cannot do "git clone", cannot do "pip install" if the wheel is absent from our wheelhouse). You may also have problems if your code performs downloads or uploads (e.g. in machine learning where downloading data from the code is often done).
JupyterHub for universities and schools
- The Pacific Institute for the Mathematical Sciences in collaboration with the Alliance and Cybera offer cloud-based hubs to universities and schools. Each institution can have its own hub where users authenticate with their credentials from that institution. The hubs are hosted on Alliance clouds and are essentially for training purposes. Institutions interested in obtaining their own hub can visit Syzygy.
Server options
Once logged in, depending on the configuration of JupyterHub, the user's web browser is redirected to either a) a previously launched Jupyter server, b) a new Jupyter server with default options, or c) a form that allows a user to set different options for their Jupyter server before pressing the Start button. In all cases, it is equivalent to accessing requested resources via an interactive job on the corresponding cluster.
Important: On each cluster, only one interactive job at a time gets a priority increase in order to start in a few seconds or minutes. That includes salloc, srun and JupyterHub jobs. If you already have another interactive job running on the cluster hosting JupyterHub, your new Jupyter session may never start before the time limit of 5 minutes.
Compute resources
For example, Server Options available on Béluga's JupyterHub are:
- Account to be used: any
def-*,rrg-*,rpp-*orctb-*account a user has access to - Time (hours) required for the session
- Number of (CPU) cores that will be reserved on a single node
- Memory (MB) limit for the entire session
- (Optional) GPU configuration: at least one GPU
- User interface (see below)
User interface
While JupyterHub allows each user to use one Jupyter server at a time on each hub, there can be multiple options under User interface:
- Jupyter Notebook (classic interface): Even though it offers many functionalities, the community is moving towards JupyterLab, which is a better platform that offers many more features.
- JupyterLab (modern interface): This is the most recommended Jupyter user interface for interactive prototyping and data visualization.
- Terminal (for a single terminal only): It gives access to a terminal connected to a remote account, which is comparable to connecting to a server through an SSH connection.
Note: JupyterHub could also have been configured to force a specific user interface. This is usually done for special events.
JupyterLab
JupyterLab is now the recommended general-purpose user interface to use on a JupyterHub. From a JupyterLab server, you can manage your remote files and folders, and you can launch Jupyter applications like a terminal, (Python 3) notebooks, RStudio and a Linux desktop.
The JupyterLab interface
When JupyterLab is ready to be used, the interface has multiple panels.
Menu bar on top
- In the File menu:
- Hub Control Panel: if you want to manually stop the JupyterLab server and the corresponding job on the cluster. This is useful when you want to start a new JupyterLab server with more or less resources.
- Log Out: the JupyterHub session will end, which will also stop the JupyterLab server and the corresponding job on the cluster.
- Most other menu items are related to notebooks and Jupyter applications.
Tool selector on left
- File Browser (folder icon):
- This is where you can browse in your home, project and scratch spaces.
- It is also possible to upload files.
- Running Terminals and Kernels (stop icon):
- To stop kernel sessions and terminal sessions
- Commands
- Property Inspector
- Open Tabs:
- To navigate between application tabs.
- To close application tabs (the corresponding kernels remain active).
- Software (blue diamond sign):
- Alliance modules can be loaded and unloaded in the JupyterLab session. Depending on the modules loaded, icons directing to the corresponding Jupyter applications will appear in the Launcher tab.
- The search box can search for any available module and show the result in the Available Modules subpanel. Note: Some modules are hidden until their dependency is loaded: we recommend that you first look for a specific module with
module spider module_namefrom a terminal. - The next subpanel is the list of Loaded Modules in the whole JupyterLab session. Note: While
pythonandipython-kernelmodules are loaded by default, additional modules must be loaded before launching some other applications or notebooks. For example:scipy-stack. - The last subpanel is the list of Available modules, similar to the output of
module avail. By clicking on a module's name, detailed information about the module is displayed. By clicking on the Load link, the module will be loaded and added to the Loaded Modules list.
Applications area on right
- The Launcher tab is open by default.
- It contains all available Jupyter applications and notebooks, depending on which modules are loaded
Status bar at the bottom
- By clicking on the icons, this brings you to the Running Terminals and Kernels tool.
Prebuilt applications
JupyterLab offers access to a terminal, an IDE (Desktop), a Python console and different options to create text and markdown files. This section presents only the main supported Jupyter applications that work with our software stack.
Command line interpreters
Julia console
To enable the Julia 1.x console launcher, an ijulia-kernel module needs to be loaded. When launched, a Julia interpreter is presented in a new JupyterLab tab.
Python console
The Python 3.x console launcher is available by default in a new JupyterLab session. When launched, a Python 3 interpreter is presented in a new JupyterLab tab.
Terminal
This application launcher will open a terminal in a new JupyterLab tab:
- The terminal runs a (Bash) shell on the remote compute node without the need of an SSH connection.
- Gives access to the remote filesystems (
/home,/project,/scratch). - Allows running compute tasks.
- Gives access to the remote filesystems (
- The terminal allows copy-and-paste operations of text:
- Copy operation: select the text, then press Ctrl+C.
- Note: Usually, Ctrl+C is used to send a SIGINT signal to a running process, or to cancel the current command. To get this behaviour in JupyterLab's terminal, click on the terminal to deselect any text before pressing Ctrl+C.
- Paste operation: press Ctrl+V.
- Copy operation: select the text, then press Ctrl+C.
Available notebook kernels
Julia notebook
To enable the Julia 1.x notebook launcher, an ijulia-kernel module needs to be loaded. When launched, a Julia notebook is presented in a new JupyterLab tab.
Python notebook
If any of the following scientific Python packages is required by your notebook, before you open this notebook, you must load the scipy-stack module from the JupyterLab Softwares tool:
ipython,ipython_genutils,ipykernel,ipyparallelmatplotlibnumpypandasscipy- Other notable packages are
Cycler,futures,jupyter_client,jupyter_core,mpmath,pathlib2,pexpect,pickleshare,ptyprocess,pyzmq,simplegeneric,sympy,tornado,traitlets. - And many more (click on the
scipy-stackmodule to see all Included extensions).
Note: You may also install needed packages by running for example the following command inside a cell: !pip install --no-index numpy.
- For some packages (like
plotly, for example), you may need to restart the notebook's kernel before importing the package. - The installation of packages in the default Python kernel environment is temporary to the lifetime of the JupyterLab session; you will have to reinstall these packages the next time you start a new JupyterLab session. For a persistent Python environment, you must configure a custom Python kernel.
To open an existing Python notebook:
- Go back to the File Browser.
- Browse to the location of the
*.ipynbfile. - Double-click on the
*.ipynbfile.- This will open the Python notebook in a new JupyterLab tab.
- An IPython kernel will start running in the background for this notebook.
To open a new Python notebook in the current File Browser directory:
- Click on the Python 3.x launcher under the Notebook section.
- This will open a new Python 3 notebook in a new JupyterLab tab.
- A new IPython kernel will start running in the background for this notebook.
Other applications
OpenRefine
To enable the OpenRefine application launcher, an openrefine module needs to be loaded. Depending on the software environment version, the latest version of OpenRefine should be loaded:
- with
StdEnv/2023, no OpenRefine module is available as of August 2024; please loadStdEnv/2020first. - with
StdEnv/2020, load module:openrefine/3.4.1.
This OpenRefine launcher will open or reopen an OpenRefine interface in a new web browser tab.
- It is possible to reopen an active OpenRefine session after the web browser tab was closed.
- The OpenRefine session will end when the JupyterLab session ends.
RStudio
To enable the RStudio application launcher, load the module: rstudio-server/4.3
This RStudio launcher will open or reopen an RStudio interface in a new web browser tab.
- It is possible to reopen an active RStudio session after the web browser tab was closed.
- The RStudio session will end when the JupyterLab session ends.
VS Code
To enable the VS Code (Visual Studio Code) application launcher, a code-server module needs to be loaded. Depending on the software environment version, the latest version of VS Code should be loaded:
- with
StdEnv/2023, load module:code-server/4.92.2. - with
StdEnv/2020, load module:code-server/3.12.0.
This VS Code launcher will open or reopen the VS Code interface in a new web browser tab.
- For a new session, the VS Code session can take up to 3 minutes to complete its startup.
- It is possible to reopen an active VS Code session after the web browser tab was closed.
- The VS Code session will end when the JupyterLab session ends.
Desktop
This Desktop launcher will open or reopen a remote Linux desktop interface in a new web browser tab:
- This is equivalent to running a VNC server on a compute node, then creating an SSH tunnel and finally using a VNC client, but you need nothing of all this with JupyterLab!
- It is possible to reopen an active desktop session after the web browser tab was closed.
- The desktop session will end when the JupyterLab session ends.
Running notebooks as Python scripts
1. From the console, or in a new notebook cell, install nbconvert :
!pip install --no-index nbconvert
2. Convert your notebooks to Python scripts
!jupyter nbconvert --to python my-current-notebook.ipynb
3. Create your non-interactive submission script, and submit it.
In your submission script, run your converted notebook with:
python my-current-notebook.py
And submit your non-interactive job:
[name@server ~]$ sbatch my-submit.sh
Possible error messages
Most JupyterHub errors are caused by the underlying job scheduler which is either unresponsive or not able to find appropriate resources for your session. For example:
Spawn failed: Timeout
- When starting a new session, JupyterHub automatically submits on your behalf a new interactive job to the cluster. If the job does not start within five minutes, a "Timeout" error message is raised and the session is cancelled.
- Just like any interactive job on any cluster, a longer requested time can cause a longer wait time in the queue. Requesting a GPU or too many CPU cores can also cause a longer wait time. Make sure to request only the resources you need for your session.
- If you already have another interactive job on the same cluster, your Jupyter session will be waiting along with other regular batch jobs in the queue. If possible, stop or cancel any other interactive job before using JupyterHub.
- There may be just no resource available at the moment. Check the status page for any issue and try again later.