META-Farm
Overview[edit]
META is a suite of scripts designed in SHARCNET to automate high-throughput computing, that is, running a large number of related calculations. This practice is sometimes called "farming", "serial farming", or "task farming". META works on all Alliance national systems, and could also be used on other clusters which use the same setup (most importantly, which use the Slurm scheduler).
We will use the term "case" in this article to describe one independent computation. In contrast, a "job" will mean an invocation of the job scheduler (Slurm). A "case" may involve the execution of a serial program, a parallel program, or a GPU-using program. A job might handle several cases.
META has the following features:
- Two modes of operation:
- SIMPLE mode, which handles one case per job.
- META node, which handles many cases per job.
- Dynamic workload balancing in META mode.
- Capture the exit status of all individual cases.
- Automatically resubmit all the cases which failed or never ran.
- Submit and independently operate multiple "farms" (groups of cases) on the same cluster.
- Can automatically run a post-processing job once all the cases have been processed successfully.
Some technical requirements:
- Each case to be computed must be described as a separate line in a file table.dat.
- One can run multiple farms independently, but each farm must have its own directory.
In the META mode, the number of actual jobs (so-called "meta-jobs") submitted by the package is usually much smaller than the number of cases to process. Each meta-job can process multiple lines from table.dat (multiple cases). A collection of meta-jobs will read lines from table.dat, starting from the first line, in a serialized manner using the lockfile mechanism to prevent a race condition. This ensures a good dynamic workload balance between meta-jobs, as meta-jobs which happen to handle shorter cases will process more of them.
Not all meta-jobs need to ever run in the META mode. The first meta-job to run will start processing lines from table.dat; if and when the second job starts, it joins the first one, and so on. If the run-time of an individual meta-job is long enough, all the cases might be processed with just a single meta-job.
META vs. GLOST[edit]
There are three important advantages of the META package over other approaches like GLOST where farm processing is done by bundling up all the jobs into a large parallel (MPI) job:
- As the scheduler has full flexibility to start individual meta-jobs when it wants, the queue wait time can be dramatically shorter with the META package than with GLOST. Consider a large farm where 1000 CPU cores need to be used for 3 days. With META, some meta-jobs start to run and produce the first results within minutes. With GLOST, with a 1000-way MPI job, queue wait time can be weeks, so it'll be weeks before you see your very first result.
- With GLOST, at the end of the farm computations, some MPI ranks will finish earlier and will sit idle until the very last -- the slowest -- MPI rank ends. In META package there is no such waste at the end of the farm -- individual meta-jobs exit earlier if they have no more workload to process.
- GLOST and other similar packages do not support automated resubmission of the cases which failed or never ran. META has this feature, and it is very easy to use.
The META webinar[edit]
A webinar was recorded on October 6th, 2021 describing the META package. You can view it here.
Quick start[edit]
If you are impatient to start using the package, just follow the steps listed below. But it is highly recommended to also read the rest of the page.
- Log in to a cluster.
- Load the
meta-farmmodule:
$ module load meta-farm
- Choose a name for a farm directory, e.g.
Farm_name, and create it with the following command:
$ farm_init.run Farm_name
- The above command will also create a few important files inside the farm directory, some of which you will need to customize.
- Copy your executable and input files to the farm directory. (You may skip this step if you plan to use full paths everywhere.)
- Edit the file
table.datinside the farm directory. This is a text file describing one case (one independent computation) per line. For examples, see one or more of:- single_case.sh
- Example: Numbered input files (advanced)
- Example: Input file must have the same name (advanced)
- Using all the columns in the cases table explicitly (advanced)
- Modify
single_case.shscript if needed. In many cases you don't have to make any changes. For more information see one or more of: - Modify
job_script.shfile to suit your needs as described at job_script.sh below. In particular, use a correct account name, and set an appropriate job run-time. For more about run-times, see Estimating the run-time and number of meta-jobs. - Inside the farm directory, execute
$ submit.run -1
for the one case per job (SIMPLE) mode, or
$ submit.run N
for the many cases per job (META) mode, where N is the number of meta-jobs to use. N should be significantly smaller than the total number of cases.
To run another farm concurrently with the first one, run farm_init.run again (providing a different farm name) and customize the files single_case.sh and job_script.sh inside the new farm directory, then create a new table.dat file there. Also copy the executable and all the input files as needed. Now you can execute the submit.run command inside the second farm directory to submit the second farm.
List of commands[edit]
- farm_init.run : Initialize a farm. See Quick start above.
- submit.run : Submit the farm to the scheduler. See submit.run below.
- resubmit.run : Resubmit all computations which failed or never ran as a new farm. See Resubmitting failed cases below.
- list.run List all the jobs with their current state for the farm.
- query.run Provide a short summary of the state of the farm, showing the number of queued, running, and completed jobs. More convenient than using
list.runwhen the number of jobs is large. It will also print the progress--- that is, the number of processed cases vs. the total number of cases--- both for the current run, and globally. - kill.run: Kill all the running and queued jobs in the farm.
- prune.run: Remove only queued jobs.
- Status.run (capital "S"!) List statuses of all processed cases. With the optional
-f, the non-zero status lines (if any) will be listed at the end. - clean.run: Delete all the files in the farm directory (including subdirectories if any present), except for
job_script.sh, single_case.sh, final.sh, resubmit_script.sh, config.h,andtable.dat. It will also delete all files associated with this farm in the/home/$USER/tmpdirectory. Be very careful with this script!
All of these commands (except for farm_init.run itself) have to be executed inside a farm directory, that is, a directory created by farm_init.run.
Small number of cases (SIMPLE mode)[edit]
Recall that a single execution of your code is a "case" and a "job" is an invocation of the Slurm scheduler. If:
- the total number of cases is fairly small--- say, less than 500, and
- each case runs for at least 10 minutes,
then it is reasonable to dedicate a separate job to each case using SIMPLE mode. Otherwise you should consider using META mode to handle many cases per job, for which please see Large number of cases (META mode) below.
The three essential scripts are the command submit.run, and two user-customizable scripts single_case.sh and job_script.sh.
submit.run[edit]
The command submit.run has one obligatory argument, the number of jobs to submit, N:
$ submit.run N [-auto] [optional_sbatch_arguments]
If N=-1, you are requesting the SIMPLE mode ("submit as many jobs as there are lines in table.dat"). If N is a positive integer, you are requesting the META mode (multiple cases per job), with N being the number of meta-jobs requested. Any other value for N is an error.
If the optional switch -auto is present, the farm will resubmit itself automatically at the end, more than once if necessary, until all the cases from table.dat have been processed. This feature is described at Running resubmit.run automatically.
If a file named final.sh is present in the farm directory, submit.run will treat it as a job script for a post-processing job and it will be launched automatically once all the cases from table.dat have been successfully processed. See Running a post-processing job automatically for more details.
If you supply any other arguments, they will be passed on to the Slurm command sbatch used to launch all meta-jobs for this farm.
single_case.sh[edit]
The function of single_case.sh is to read one line from table.dat, parse it, and use the contents of that line to launch your code for one case.
You may wish to customize single_case.sh for your purposes.
The version of single_case.sh provided by farm_init.run treats each line in table.dat as a literal command and executes it in its own subdirectory RUNyyy, where yyy is the case number. Here is the relevant section of single_case.sh:
...
# ++++++++++++++++++++++ This part can be customized: ++++++++++++++++++++++++
# Here:
# $ID contains the case id from the original table (can be used to provide a unique seed to the code etc)
# $COMM is the line corresponding to the case $ID in the original table, without the ID field
# $METAJOB_ID is the jobid for the current meta-job (convenient for creating per-job files)
mkdir -p RUN$ID
cd RUN$ID
echo "Case $ID:"
# Executing the command (a line from table.dat)
# It's allowed to use more than one shell command (separated by semicolons) on a single line
eval "$COMM"
# Exit status of the code:
STATUS=$?
cd ..
# +++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
...
Consequently, if you are using the unmodified single_case.sh then each line of table.dat should contain a complete command.
This may be a compound command, that is, several commands separated by semicolons (;).
Typically table.dat will contain a list of identical commands differentiated only by their arguments, but it need not be so.
Any executable statement can go into table.dat.
Your table.dat could look like this:
/home/user/bin/code1 1.0 10 2.1 cp -f ~/input_dir/input1 .; ~/code_dir/code ./code2 < IC.2
If you intend to execute the same command for every case and don't wish to repeat it on every line of table.dat,
then you can edit single_case.sh to include the common command.
Then edit your table.dat to contain only the arguments and/or redirects for each case.
For example, here is a modification of single_case.sh which includes the command
(/path/to/your/code), takes the contents of table.dat as arguments to
that command, and uses the case number $ID as an additional argument:
- single_case.sh:
...
# ++++++++++++++++++++++ This part can be customized: ++++++++++++++++++++++++
# Here we use $ID (case number) as a unique seed for Monte-Carlo type serial farming:
/path/to/your/code -par $COMM -seed $ID
STATUS=$?
# +++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
...
- table.dat:
12.56
21.35
...
Note: If your code doesn't need to read any arguments from table.dat, you still have to generate table.dat, with the number of lines equal to the number of cases you want to compute. In this case, it doesn't matter what you put inside table.dat -- all that matters is the total number of the lines. The key line in the above example might then look like
/path/to/your/code -seed $ID
Another note: You do not need to insert line numbers at the beginning of each line of table.dat. The script submit.run will modify table.dat to add line numbers if it doesn't find them there.
STATUS and handling errors[edit]
What is STATUS for in single_case.sh? It is a variable which should be set to “0” if your case was computed correctly, and some positive value (that is, greater than 0) otherwise. It is very important: It is used by resubmit.run to figure out which cases failed so they can be re-computed. In the provided version of single_case.sh, STATUS is set to the exit code of your program. This may not cover all potential problems, since some programs produce an exit code of zero even if something goes wrong. You can change how STATUS is set by editing single_case.sh.
For example if your code is supposed to write a file (say, out.dat) at the end of each case, test whether the file exists and set STATUS appropriately.
In the following code fragment, $STATUS will be positive if either the exit code from the program is positive, or if out.dat doesn't exist or is empty:
STATUS=$?
if test ! -s out.dat
then
STATUS=1
fi
job_script.sh[edit]
The file job_script.sh is the job script which will be submitted to SLURM for all meta-jobs in your farm.
Here is the default version created for you by farm_init.run:
#!/bin/bash
# Here you should provide the sbatch arguments to be used in all jobs in this serial farm
# It has to contain the runtime switch (either -t or --time):
#SBATCH -t 0-00:10
#SBATCH --mem=4G
#SBATCH -A Your_account_name
# Don't change this line:
task.run
At the very least you should change the account name (the -A switch), and the meta-job run-time (the -t switch).
In SIMPLE mode, you should set the run-time to be somewhat longer than the longest expected individual case.
Important: Your job_script.sh must include the run-time switch (either -t or -time).
This cannot be passed to sbatch as an optional argument to submit.run.
Sometimes the following problem happens: A meta-job may be allocated to a node which has a defect, thereby causing your program to fail instantly. For example, perhaps your program needs a GPU but the GPU you're assigned is malfunctioning, or perhaps the /project file system is not mounted. (Please report such a defective node to support@tech.alliancecan.ca if you detect one!) But when it happens, that single bad meta-job can quickly churn through table.dat, so your whole farm fails. If you can anticipate such problems, you can add tests to job_script.sh before the task.run line. For example, the following modification will test for the presence of an NVidia GPU, and if none is found it will force the meta-job to exit before it starts failing your cases:
nvidia-smi >/dev/null
retVal=$?
if [ $retVal -ne 0 ]; then
exit 1
fi
task.run
There is a utility gpu_test which does a similar job to nvidia_smi in the above example.
On Graham, Cedar, or Beluga you can copy it to your ~/bin directory:
cp ~syam/bin/gpu_test ~/bin
The META package has a built-in mechanism which tries to detect problems of this kind and kill a meta-job which churns through the cases too quickly. The two relevant parameters, N_failed_max and dt_failed are set in the file config.h. The protection mechanism is triggered when the first $N_failed_max cases are very short - less than $dt_failed seconds in duration. The default values are 5 and 5, so by default a meta-job will stop if the first 5 cases all finish in less than 5 seconds. If you get false triggering of this protective mechanism because some of your normal cases have run-time shorter than $dt_failed, reduce the value of dt_failed in config.h.
Output files[edit]
Once one or more meta-jobs in your farm are running, the following files will be created in the farm directory:
OUTPUT/slurm-$JOBID.out, one file per meta-job: Standard output from meta-jobs.STATUSES/status.$JOBID, one file per meta-job: Files containing the statuses of the processed cases.
In both cases, $JOBID stands for the jobid of the corresponding meta-job.
One more directory, MISC, will also be created inside the root farm directory. It contains some auxiliary data.
Also, every time submit.run is run it will create a unique subdirectory inside /home/$USER/tmp.
Inside that subdirectory, some small scratch files will be created, such as files used by lockfile to serialize certain operations inside the jobs.
These subdirectories have names $NODE.$PID, where $NODE is the name of the current node (typically a login node), and $PID is the unique process ID for the script.
Once the farm execution is done, one can safely erase this subdirectory.
This will happen automatically if you run clean.run, but be careful! clean.run also deletes all the results produced by your farm!
Resubmitting failed cases[edit]
The command resubmit.run takes the same arguments as submit.run:
$ resubmit.run N [-auto] [optional_sbatch_arguments]
resubmit.run:
- analyzes all those
status.*files (see Output files above); - figures out which cases failed and which never ran for whatever reason (e.g. because of the meta-jobs' run-time limit);
- creates a new case table (adding “_” at the end of the original table name), which lists only the cases which still need to be run;
- launches a new farm for those cases.
You cannot run resubmit.run until all the jobs from the original run are done or killed.
If some cases still fail or do not run, one can resubmit the farm as many times as needed. Of course, if certain cases fail repeatedly then there must a be a problem with either the program you are running or its input. In this case you may wish to use the command Status.run (capital S!) which displays the statuses for all computed cases. With the optional argument -f, Status.run will sort the output according to the exit status, showing cases with non-zero status at the bottom, to make them easier to spot.
Similarly to submit.run, if the optional switch -auto is present the farm will resubmit itself automatically at the end, more than once if necessary. This advanced feature is described at Resubmitting failed cases automatically.
Large number of cases (META mode)[edit]
META mode overview[edit]
The SIMPLE (one case per job) mode works fine when the number of cases is fairly small (<500). When the number of cases is much greater than 500, the following problems may arise:
- Each cluster has a limit on how many jobs a user can have at one time. (E.g. for Graham, it is 1000.)
- With a very large number of cases, each case computation is typically short. If one case runs for <20 min, CPU cycles may be wasted due to scheduling overheads.
META mode is the solution to these problems.
Instead of submitting a separate job for each case, a smaller number of "meta-jobs" are submitted, each of which processes multiple cases.
To enable META mode the first argument to submit.runt should be the desired number of meta-jobs,
which should be a fairly small number-- much smaller than the number of cases to process. E.g.:
$ submit.run 32
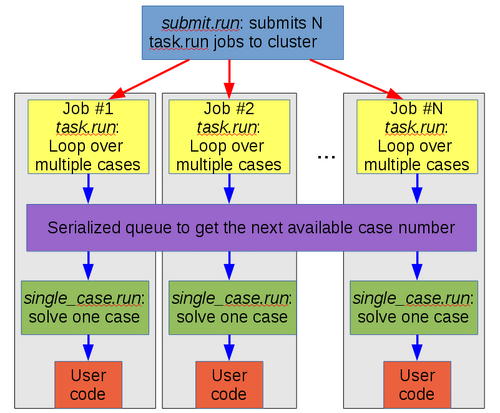
Since each case may take a different amount of time to process, META modes uses a dynamic workload-balancing scheme. This is how META mode is implemented:
As the above diagram shows, each job executes the same script, task.run. Inside that script, there is a while loop for the cases. Each iteration of the loop has to go through a serialized portion of the code (that is, only one job at a time can be in that section of code), where it gets the next case to process from table.dat. Then the script single_case.sh (see section #single_case.sh script) is executed once for each case, which in turn calls the user code.
This approach results in dynamic workload balancing achieved across all the running "meta-jobs" belonging to the same farm. The algorithm is illustrated by the diagram below:
This can be seen more clearly in this animation from the META webinar.
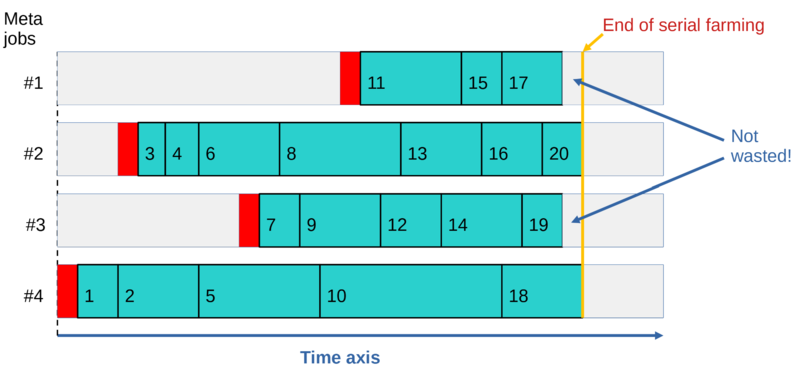
The dynamic workload balancing results in all meta-jobs finishing around the same time, regardless of how different the run-times are for individual cases, regardless of how fast CPUs are on different nodes, and regardless of when individual "meta-jobs" start. In addition, not all meta-jobs need to start running for all the cases to be processed, and if a meta-job dies (e.g. due to a node crash), at most one case will be lost. The latter can be easily rectified with resubmit.run; see #Resubmitting failed/never-run jobs.
Not all of the requested meta-jobs will necessarily run, depending on how busy the cluster is. But as described above, in META mode you will eventually get all your results regardless of how many meta-jobs run, although you might need to use resubmit.run to complete a particularly large farm.
Estimating the run-time and number of meta-jobs[edit]
How should you figure out the optimum number of meta-jobs, and the run-time to be used in job_script.sh?
First you need to figure out the average run-time for an individual case (a single line in table.dat). Supposing your application program is not parallel, allocate a single CPU core with salloc, then execute single_case.sh there for a few different cases. Measure the total run-time and divide that by the number of cases you ran to get an estimate of the average case run-time. This can be done with a shell for loop:
$ N=10; time for ((i=1; i<=$N; i++)); do ./single_case.sh table.dat $i ; done
Divide the "real" time output by the above command by $N to get the average case run-time estimate. Let's call it dt_case.
Estimate the total CPU time needed to process the whole farm by multiplying dt_case by the number of cases, that is, the number of lines in table.dat.
If this is in CPU-seconds, dividing that by 3600 gives you the total number of CPU-hours.
Multiply that by something like 1.1 or 1.3 to have a bit of a safety margin.
Now you can make a sensible choice for the run-time of meta-jobs, and that will also determine the number of meta-jobs needed to finish the whole farm.
The run-time you choose should be significantly larger than the average run-time of an individual case, ideally by a factor of 100 or more. It must definitely be larger than the longest run-time you expect for an individual case. On the other hand it should not be too large; say, no more than 3 days. The longer a job's run-time is, the longer it will usually wait to be scheduled. On Alliance general-purpose clusters, a good choice would be 12h or 24h due to scheduling policies. Once you have settled on a run-time, divide the total number of CPU-hours by the run-time you have chosen (in hours) to get the required number of meta-jobs. Round up this number to the next integer.
With the above choices, the queue wait time should be fairly small, and the throughput and efficiency of the farm should be fairly high.
Let's consider a specific example. Suppose you ran the above for loop on a dedicated CPU obtained with salloc, and the output said the "real" time was 15m50s, which is 950 seconds. Divide that by the number of sample cases, 10, to find that the average time for an individual case is 95 seconds. Suppose also the total number of cases you have to process (the number of lines in table.dat) is 1000. The total CPU time required to compute all your cases is then 95 x 1000 = 95,000 CPU-seconds = 26.4 CPU-hours. Multiply that by a factor of 1.2 as a safety measure, to yield 31.7 CPU-hours. A run-time of 3 hours for your meta-jobs would work here, and should lead to good queue wait times. Edit the value of the #SBATCH -t in job_script.sh to be 3:00:00. Now estimate how many meta-jobs you'll need to process all the cases: N = 31.7 core-hours / 3 hours = 10.6, which rounded up to the next integer is 11. Then you can launch the farm by executing a single submit.run 11.
If the number of jobs in the above analysis is larger than 1000, you have a particularly large farm. The maximum number of jobs which can be submitted on Graham and Beluga is 1000, so you won't be able to run the whole collection with a single command. The workaround would be to go through the following sequence of commands. Remember each command can only be executed after the previous farm has finished running:
$ submit.run 1000
$ resubmit.run 1000
$ resubmit.run 1000
...
If this seems rather tedious, consider using an advanced feature of the META package for such large farms: Resubmitting failed cases automatically. This will fully automate the farm resubmission steps.
Reducing waste[edit]
Here is one potential problem when one is running multiple cases per job: What if the number of running meta-jobs times the requested run-time per meta-job (say, 3 days) is not enough to process all your cases? E.g., you managed to start the maximum allowed 1000 meta-jobs, each of which has a 3-day run-time limit. That means that your farm can only process all the cases in a single run if the average_case_run_time x N_cases < 1000 x 3d = 3000 CPU days. Once your meta-jobs start hitting the 3-day run-time limit, they will start dying in the middle of processing one of your cases. This will result in up to 1000 interrupted cases calculations. This is not a big deal in terms of completing the work--- resubmit.run will find all the cases which failed or never ran, and will restart them automatically. But this can become a waste of CPU cycles. On average, you will be wasting 0.5 x N_jobs x average_case_run_time. E.g., if your cases have an average run-time of 1 hour, and you have 1000 meta-jobs running, you will waste about 500 CPU-hours or about 20 CPU-days, which is not acceptable.
Fortunately, the scripts we are providing have some built-in intelligence to mitigate this problem. This is implemented in task.run as follows:
- The script measures the run-time of each case, and adds the value as one line in a scratch file
timescreated in directory/home/$USER/tmp/$NODE.$PID/. (See Output files.) This is done by all running meta-jobs. - Once the first 8 cases were computed, one of the meta-jobs will read the contents of the file
timesand compute the largest 12.5% quantile for the current distribution of case run-times. This will serve as a conservative estimate of the run-time for your individual cases, dt_cutoff. The current estimate is stored in filedt_cutoffin/home/$USER/tmp/$NODE.$PID/. - From now on, each meta-job will estimate if it has the time to finish the case it is about to start computing, by ensuring that t_finish - t_now > dt_cutoff. Here, t_finish is the time when the job will die because of the job's run-time limit, and t_now is the current time. If it computes that it doesn't have the time, it will exit early, which will minimize the chance of a case aborting half-way due to the job's run-time limit.
- At every subsequent power of two number of computed cases (8, then 16, then 32 and so on) dt_cutoff is recomputed using the above algorithm. This will make the dt_cutoff estimate more and more accurate. Power of two is used to minimize the overheads related to computing dt_cutoff; the algorithm will be equally efficient for both very small (tens) and very large (many thousands) number of cases.
- The above algorithm reduces the amount of CPU cycles wasted due to jobs hitting the run-time limit by a factor of 8, on average.
As a useful side effect, every time you run a farm you get individual run-times for all of your cases stored in /home/$USER/tmp/$NODE.$PID/times.
You can analyze that file to fine-tune your farm setup, for profiling your code, etc.
If more help is needed[edit]
See META: Advanced features and troubleshooting for more detailed discussion of some features, and for troubleshooting suggestions.
If you need more help, contact Technical support, mentioning the name of the package (META), and the name of the staff member who wrote the software (Sergey Mashchenko).
Glossary[edit]
- case: One independent computation. The file
table.datshould list one case per line. - farm / farming (verb): Running many jobs on a cluster which carry out independent (but related) computations, of the same kind.
- farm (noun): The directory and files involved in running one instance of the package.
- meta-job: A job which can process multiple cases (independent computations) from
table.dat. - META mode: The mode of operation of the package in which each job can process multiple cases from
table.dat. - SIMPLE mode: The mode of operation of the package in which each job will process only one case from
table.dat.